Basics
Summarized notes from Introduction to Data Mining (Int'l ed), Chapter 5
-
Basic clustering problem
- given: set of objects \(X\), inter-object distances \(D(x,y) = D(y,x) \enspace x,y \in X\)
- output: partition \(P_D(x) = P_D(y)\) if \(x,y\) are in the same cluster
-
divide data into meaningful and/or useful groups, to describe or summarize data
- clusters describe data instances with similar characteristics
- ideally: small distance between points in cluster; long distance between clusters
-
Some utility applications: data summarization, compression, efficiency in finding nearest neighbors
-
clustering problem is very much driven by specific choice of algorithm
Clustering techniques
-
based on nesting of clusters:
- partitional (un-nested): divides data into non-overlapping clusters such that each instance is in exactly one cluster
- hierarchical (nested): clusters are permitted to have sub-clusters; can be visualized as a tree
-
based on count of instance memberships:
- exclusive: each instance belongs to a single cluster
- overlapping / non-exclusive: instance can belong to more than one cluster
- fuzzy / soft: (probabilistic) instances membership in each cluster is determined by weigh 0-1, and typical additional constrain requires sum of weights equal to 1 => convert fuzzy to exclusive by assigning each object to cluster with highest weight
-
based on instance membership in any cluster:
- complete: each instance is assigned to some cluster
- partial: not all instances are assigned a cluster; primary motivations include omitting noise and outliers
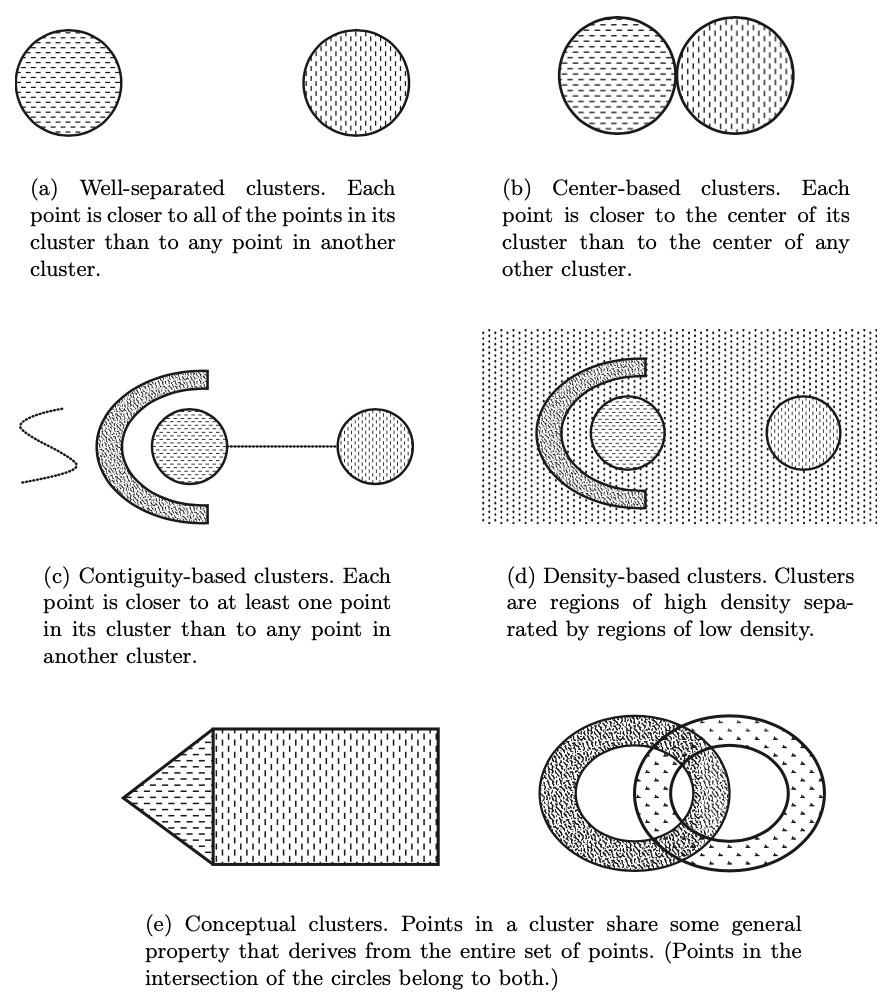
Types of clusters
-
well-separated: the distance between clusters is larger than distance between any two points within a cluster
-
prototype-based / center-based: prototype defines a cluster and each object in cluster is closer to prototype that defines the cluster than to prototype of any other cluster; the prototype may be actual record or the calculated centroid
-
contiguity-based / nearest neighbor / transitive: each point in a cluster is closer (or more similar) to one or more other points in the cluster than to any point not in the cluster.
-
graph-based: nodes represent data objects, and edges their connections; cluster is a connected component where each node is connected within cluster but there are no connections to outside the cluster => when nodes are strongly connected, then the nodes form a clique
-
density-based: cluster is dense region surrounded by a region of low density => useful strategy when clusters are irregular, intertwined or when noise or outliers are present
-
shared-property/conceptual clusters: objects in a cluster share some common property; this encompasses all of the above, but enables defining clusters not covered by previous definitions.
Clustering properties
-
Richness: for any assignment of objects to clusters there is some distance matrix \(D\) such that \(P_D\) returns that clustering \(\forall C \enspace \exists D \enspace P_D = C\)
-
Scale-invariance: scaling distances by a positive value does not change the clustering \(\forall D \enspace \forall_{K>0} \enspace P_D = P_{KD}\)
-
Consistency: shrinking intra-cluster distances and expanding inter-cluster distance does not change the clustering \(P_D = P_{D'}\)
Impossibility theorem: no clustering algorithm can have all 3 properties.
Other considerations:
- order dependence: whether results differ based on the order of processing of data points
- nondeterminism: whether different runs give different results
- scalability: how large datasets the clustering algorithm can reasonably handle
- parameter selection: number of parameters and selection of optimal parameter values
Algorithms
| Algorithm | Description | Category | Time Complexity (optimal) | Space Complexity |
|---|---|---|---|---|
| K-means | Find \(K\) clusters represented by centroids | prototype-based, partitional, complete | \(O(IKmn)\) | \(O((m + K) n)\) |
| Agglomerative hierarchical clustering | Find hierarchical clusters by merging clusters | prototype-based / graph-based, hierarchical, complete | \(O(m^2 \lg(m))\) | \(O(m^2)\) |
| DBSCAN | Cluster by density, ignoring low-density regions | Density-based, partitional, partial | \(O(m \log m)\) | \(O(m)\) |
| Fuzzy C-means | Points have weighted membership in clusters | prototype-based, partitional, fuzzy | \(O(LKI)\) | \(O(LK)\) |
where: \(m\) = number of points (records); \(n\) = number of attributes; \(I\) = number of iterations until convergence; \(K\) = number of clusters, \(L\) is the number of links
Also see:
- Expectation Maximization - statistical approach using mixture models
- Single-linkage clustering (SLC) - special case of agglomerative using single-link
- Self-organizing maps - clustering and data visualization based on neural networks
Cluster Validation
Some considerations:
- clustering tendency - distinguishing if non-random structure actually exists in the data
- determining correct number of clusters
- Evaluating how well the results of a cluster analysis fit the data without reference to external information.
- Comparing the results of a cluster analysis to externally known results, such as externally provided class labels.
- Comparing two sets of clusters to determine which is better
Defining cluster evaluation metrics is difficult, there are 3 general categories:
-
unsupervised: measure goodness without reference to external information (e.g. SSE); two subclasses:
- cohesion: compactness, tightness - how closely related objects in a cluster are
- separation: isolation - how distinct or well-separated clusters are
-
supervised: measures the extent to which the clustering structure discovered by a clustering algorithm matches some external structure (e.g. entropy of class labels compared to known class labels)
-
relative: Compares different clusterings or clusters, using either supervised or unsupervised metrics
Unsupervised
Cohesion and separation
-
for graph-based view of cohesion/separation:
- \(\displaystyle \text{cohesion}(C_i)\) = \(\sum_{x \in C_i, y \in C_i} proximity(x, y)\)
- \(\displaystyle \text{separation}(C_i, C_j)\) = \(\sum_{x \in C_i, y \in C_j} proximity(x, y)\)
-
proximity function can be similarity or dissimilarity
-
If clusters have prototype (centroid or menoid)
-
cohesion and separation can be defined as distance from prototype:
\(\displaystyle \text{cluster SSE}\) = \(\sum_i \sum_{x \in C_i} distance(c_i, x)^2\) -
Group sum of squares (SSB) measures distance between clusters
\(\displaystyle \text{Total SSB}\) = \(\sum_{i=1}^{K} m_i \times distance(c_i, c)^2\)
-
The result of cohesion/separation can be used to improve clustering by merging or splitting clusters
Silhouette Coefficient
-
Evaluation metric for evaluating points in a cluster. Steps:
-
Calculate \(a_i\), which is the average distance from object \(i\) to all other objects in its cluster
-
Calculate \(b_i\), which is the minimum average distance from object \(i\) and objects in any cluster not containing \(i\)
-
The silhouette coefficient is: \(s_i = (b_i - a_i)/max(a_i, b_i)\)
-
-
The range of coefficient is -1 to 1. Negative value is bad; as high as possible is good (obtained when \(a_i=0\)).
- Cluster S.C. can be computed by taking the average S.C. of its points
- Does not work well on arbitrary shapes.
Cophenetic Correlation Coefficient (CPCC)
Cophenetic Correlation Coefficient (CPCC) metric is for evaluating hierarchical clustering
- cophenetic distance between two objects is the proximity at which an agglomerative hierarchical clustering technique puts the objects in the same cluster for the first time
- CPCC calculates the difference between cophenetic distance and original distance matrix
Supervised
- supervised methods include knowing the real cluster data labels
- can help establish "ground truth" for clustering results
- similar metrics can be defined for precision, recall and F-measure
Entropy
-
measure the degree of clusters matching known class labels
-
For a set of clusters, \(e\):
\(\displaystyle e = \sum_{i=1}^{K} \frac{m_i}{m} e_i\)
-
For individual cluster, \(e_i\):
\(\displaystyle e_i = -\sum_{j=1}^{L} p_{ij} \lg p_{ij}\)
where:
- \(L\) is number of classes
- \(p_{ij} = m_{ij}/m_i\)
- \(m_i\) is number of objects in cluster \(i\)
- \(m_{ij}\) is number of objects of class \(j\) in cluster \(i\)
Purity
- the maximum \(p_{ij}\) for a cluster
-
overall purity of a cluster is
\(\displaystyle \sum_{i=1}^{K} \frac{m_i}{m} max_j(p_{ij})\)